A simple agent based infection model with Mesa and Bokeh
Background
Forecasting the outcome of infectious disease epidemics is now receiving much attention due to the ongoing COVID-19 pandemic. A traditional framework for infectious disease spread is the so-called SIR model, dividing a population into susceptible (S), infectious (I) and recovered/removed (R). These can be estimated over time with a set of differential equations given known transition rates between states. These in turn depend on parameters like the R0 for the infection. These equation based methods are called compartmental models. Agent-based models are a more recent advance that simulate many individual ‘agents’ in the population to achieve the same goal. The agents are heterogeneous, with multiple attributes and complexity emerges out of the aggregate behaviour of many agents combined. At least that’s my simplistic understanding. A simple example here served to help me understand how the agent-based approach works. It uses the Mesa Python library to build an SIR model and also illustrates ways of visualizing the simulation as the model is run using Bokeh.
Note: A follow up to this post using a network grid is here.
Imports
import time
import numpy as np
import pandas as pd
import pylab as plt
from mesa import Agent, Model
from mesa.time import RandomActivation
from mesa.space import MultiGrid
from mesa.datacollection import DataCollector
Building a simple model
The main idea with Mesa is to create two classes, one for the model and the other for the agents. The agent handles the behaviour of the individual being simulated such as how it can infect other neighbours in a grid or network. The model holds all the general parameters, a grid object for moving agents on and it also creates and tracks it’s agents. It’s really much more instructive to go through an example than describe. This code was made mostly using the Mesa tutorial and the Virus on network example.
We first make a Model class defining a grid, scheduler for tracking the order of agents being activated in time. Time periods are represented as steps and the agents can all move once in each step. Then the agent will decide if it can infect another according to where it is. The DataCollector class keeps track of agent information through the simulation. The grid is a MultiGrid class, which let more than one agent occupy a cell at once.
class InfectionModel(Model):
"""A model for infection spread."""
def __init__(self, N=10, width=10, height=10, ptrans=0.5,
death_rate=0.02, recovery_days=21,
recovery_sd=7):
self.num_agents = N
self.recovery_days = recovery_days
self.recovery_sd = recovery_sd
self.ptrans = ptrans
self.death_rate = death_rate
self.grid = MultiGrid(width, height, True)
self.schedule = RandomActivation(self)
self.running = True
self.dead_agents = []
# Create agents
for i in range(self.num_agents):
a = MyAgent(i, self)
self.schedule.add(a)
# Add the agent to a random grid cell
x = self.random.randrange(self.grid.width)
y = self.random.randrange(self.grid.height)
self.grid.place_agent(a, (x, y))
#make some agents infected at start
infected = np.random.choice([0,1], p=[0.98,0.02])
if infected == 1:
a.state = State.INFECTED
a.recovery_time = self.get_recovery_time()
self.datacollector = DataCollector(
agent_reporters={"State": "state"})
def get_recovery_time(self):
return int(self.random.normalvariate(self.recovery_days,self.recovery_sd))
def step(self):
self.datacollector.collect(self)
self.schedule.step()
We then create the Agent class. It has three possible states and transitions between them through the simulation. At each step the agent will move and then can carry out any operation such as infecting another agent in the same cell in the grid if the other agent is susceptible. The agent can also recover over time.
class State(enum.IntEnum):
SUSCEPTIBLE = 0
INFECTED = 1
REMOVED = 2
class MyAgent(Agent):
""" An agent in an epidemic model."""
def __init__(self, unique_id, model):
super().__init__(unique_id, model)
self.age = self.random.normalvariate(20,40)
self.state = State.SUSCEPTIBLE
self.infection_time = 0
def move(self):
"""Move the agent"""
possible_steps = self.model.grid.get_neighborhood(
self.pos,
moore=True,
include_center=False)
new_position = self.random.choice(possible_steps)
self.model.grid.move_agent(self, new_position)
def status(self):
"""Check infection status"""
if self.state == State.INFECTED:
drate = self.model.death_rate
alive = np.random.choice([0,1], p=[drate,1-drate])
if alive == 0:
self.model.schedule.remove(self)
t = self.model.schedule.time-self.infection_time
if t >= self.recovery_time:
self.state = State.REMOVED
def contact(self):
"""Find close contacts and infect"""
cellmates = self.model.grid.get_cell_list_contents([self.pos])
if len(cellmates) > 1:
for other in cellmates:
if self.random.random() > model.ptrans:
continue
if self.state is State.INFECTED and other.state is State.SUSCEPTIBLE:
other.state = State.INFECTED
other.infection_time = self.model.schedule.time
other.recovery_time = model.get_recovery_time()
def step(self):
self.status()
self.move()
self.contact()
Run the model
We can now run the model by simply iterating over the number of steps we want. The DataCollector object has stored agent variables along the way and this can be analysed to get model results. get_agent_vars_dataframe() returns a pandas DataFrame in long form of the state of each agent at each step.
model = InfectionModel(pop, 20, 20, ptrans=0.5)
for i in range(steps):
model.step()
agent_state = model.datacollector.get_agent_vars_dataframe()
View model states data
To make this data easier to plot we can convert it into wide form as follows.
def get_column_data(model):
"""pivot the model dataframe to get states count at each step"""
agent_state = model.datacollector.get_agent_vars_dataframe()
X = pd.pivot_table(agent_state.reset_index(),index='Step',columns='State',aggfunc=np.size,fill_value=0)
labels = ['Susceptible','Infected','Removed']
X.columns = labels[:len(X.columns)]
return X
The resulting table looks like this:
Step Susceptible Infected Removed
0 295 5 0
1 295 5 0
2 287 13 0
3 284 16 0
4 279 21 0
.. ... ... ... ...
95 7 0 180
96 7 0 180
97 7 0 180
98 7 0 180
99 7 0 180
Plot model states with Bokeh
This table can then be plotted to track each state. The code below makes a line plot for each column vs step.
def plot_states_bokeh(model,title=''):
"""Plot cases per country"""
X = get_column_data(model)
X = X.reset_index()
source = ColumnDataSource(X)
i=0
colors = Category10[3]
items=[]
p = figure(plot_width=600,plot_height=400,tools=[],title=title,x_range=(0,100))
for c in X.columns[1:]:
line = Line(x='Step',y=c, line_color=colors[i],line_width=3,line_alpha=.8,name=c)
glyph = p.add_glyph(source, line)
i+=1
items.append((c,[glyph]))
p.xaxis.axis_label = 'Step'
p.add_layout(Legend(location='center_right',
items=items))
p.background_fill_color = "#e1e1ea"
p.background_fill_alpha = 0.5
p.legend.label_text_font_size = "10pt"
p.title.text_font_size = "15pt"
p.toolbar.logo = None
p.sizing_mode = 'scale_height'
return p
Plot grid cell contents
We can also plot the contents of the model grid object to get an idea of the spatial changes in the state of each agent.
def grid_values(model):
"""Get grid cell states"""
agent_counts = np.zeros((model.grid.width, model.grid.height))
w=model.grid.width
df=pd.DataFrame(agent_counts)
for cell in model.grid.coord_iter():
agents, x, y = cell
c=None
for a in agents:
c = a.state
df.iloc[x,y] = c
return df
def plot_cells_bokeh(model):
agent_counts = np.zeros((model.grid.width, model.grid.height))
w=model.grid.width
df=grid_values(model)
df = pd.DataFrame(df.stack(), columns=['value']).reset_index()
columns = ['value']
x = [(i, "@%s" %i) for i in columns]
hover = HoverTool(
tooltips=x, point_policy='follow_mouse')
colors = Category10[3]
mapper = LinearColorMapper(palette=colors, low=df.value.min(), high=df.value.max())
p = figure(plot_width=500,plot_height=500, tools=[hover], x_range=(-1,w), y_range=(-1,w))
p.rect(x="level_0", y="level_1", width=1, height=1,
source=df,
fill_color={'field':'value', 'transform': mapper},
line_color='black')
p.background_fill_color = "black"
p.grid.grid_line_color = None
p.axis.axis_line_color = None
p.toolbar.logo = None
return p
Plot the model as it runs.
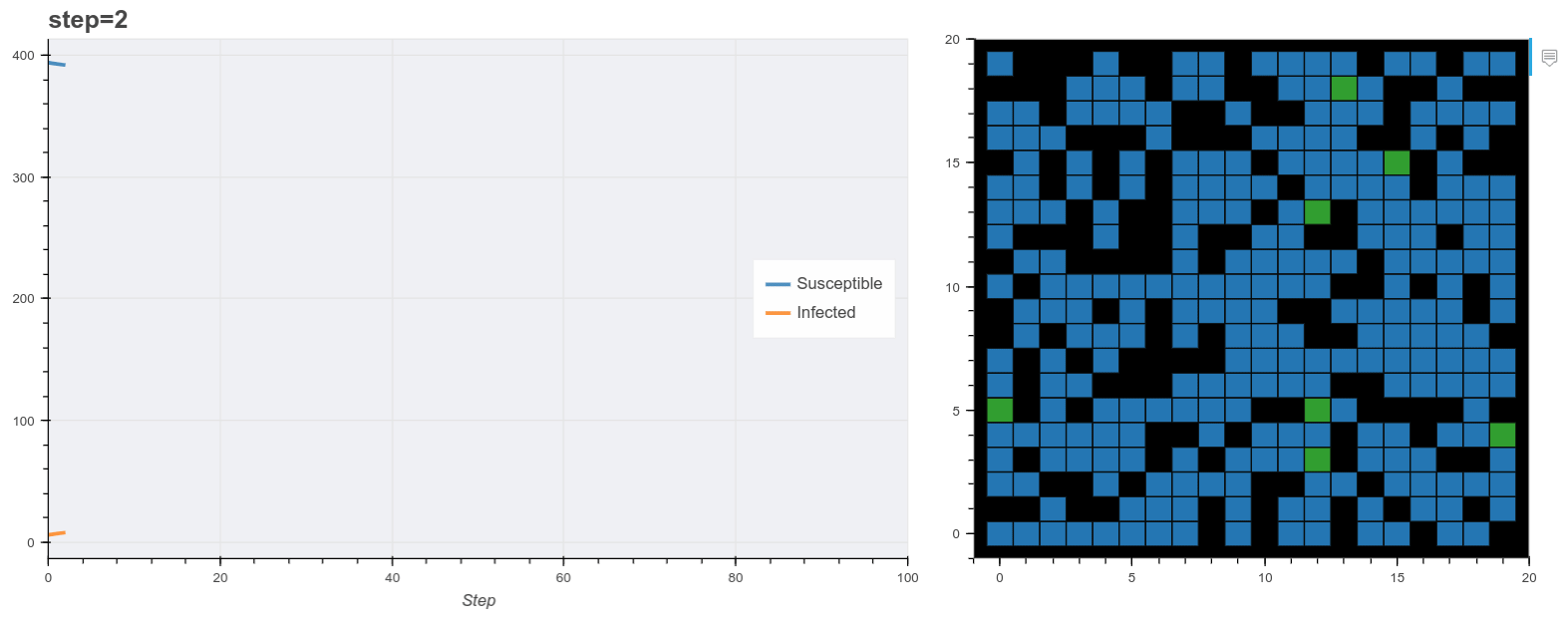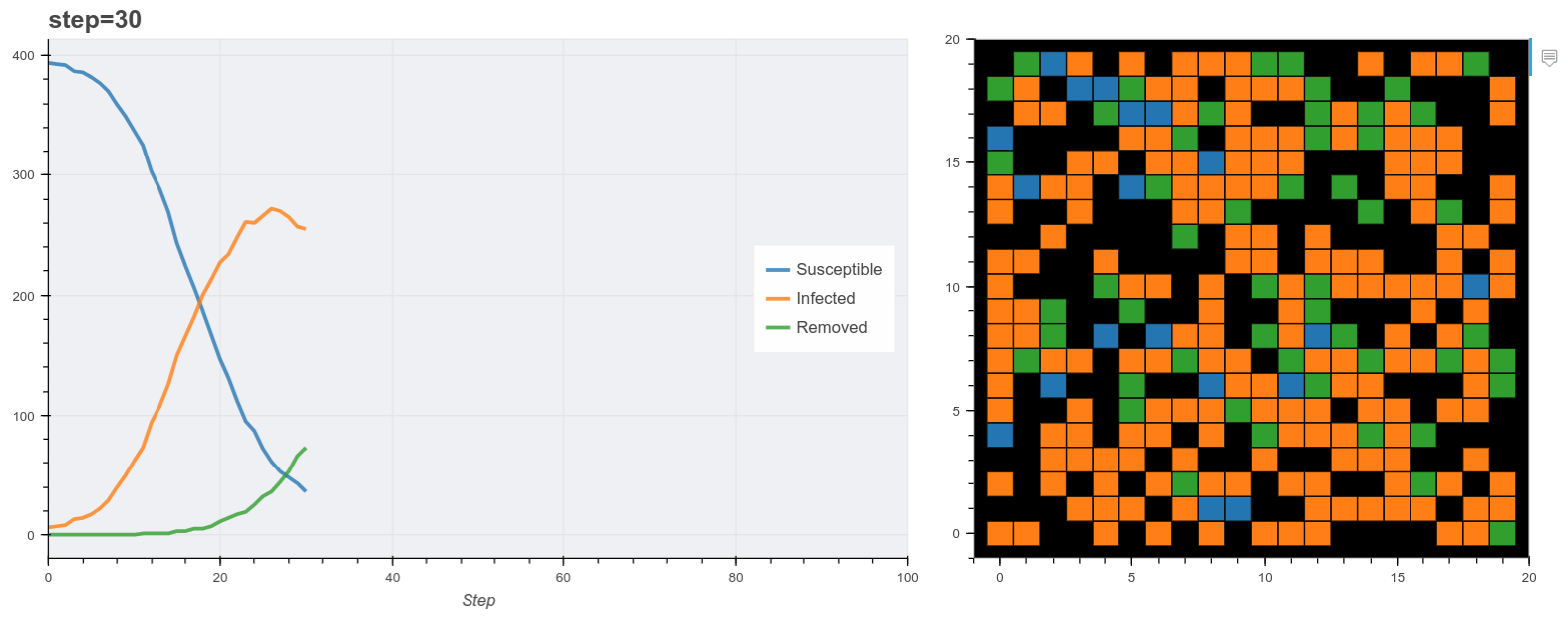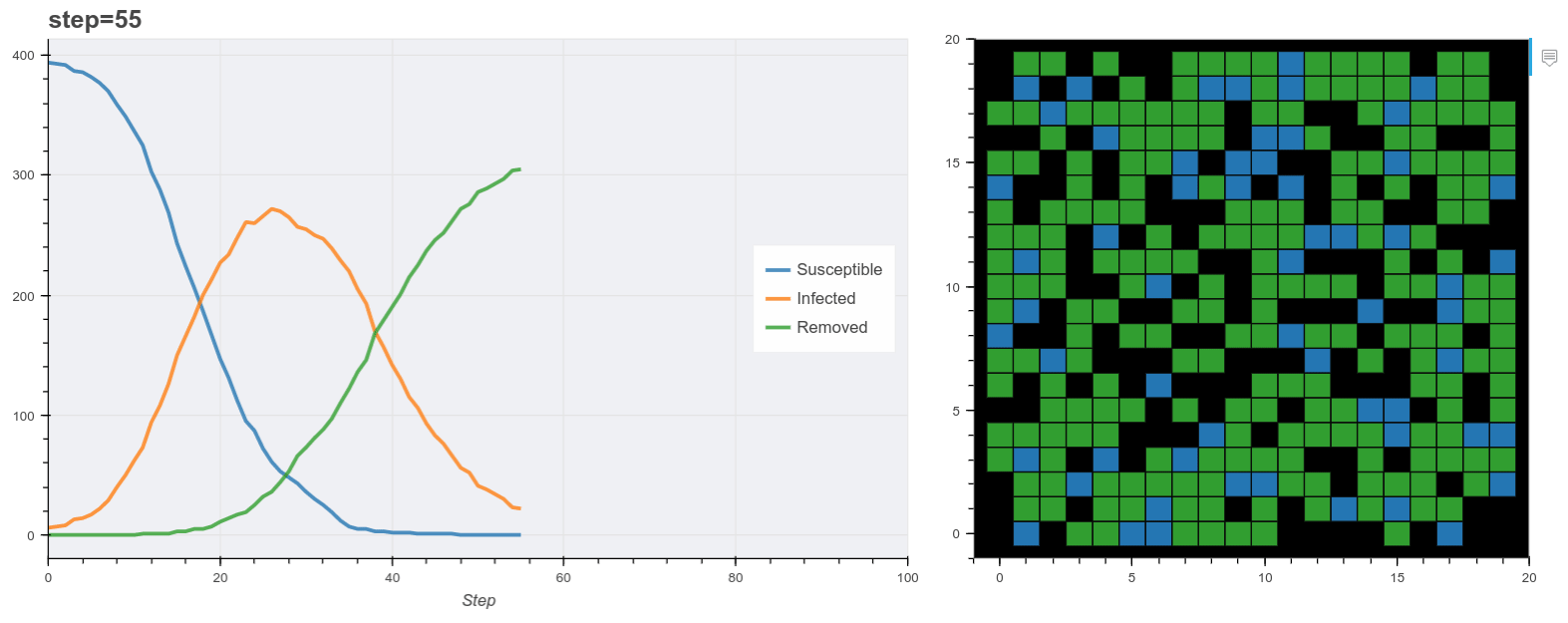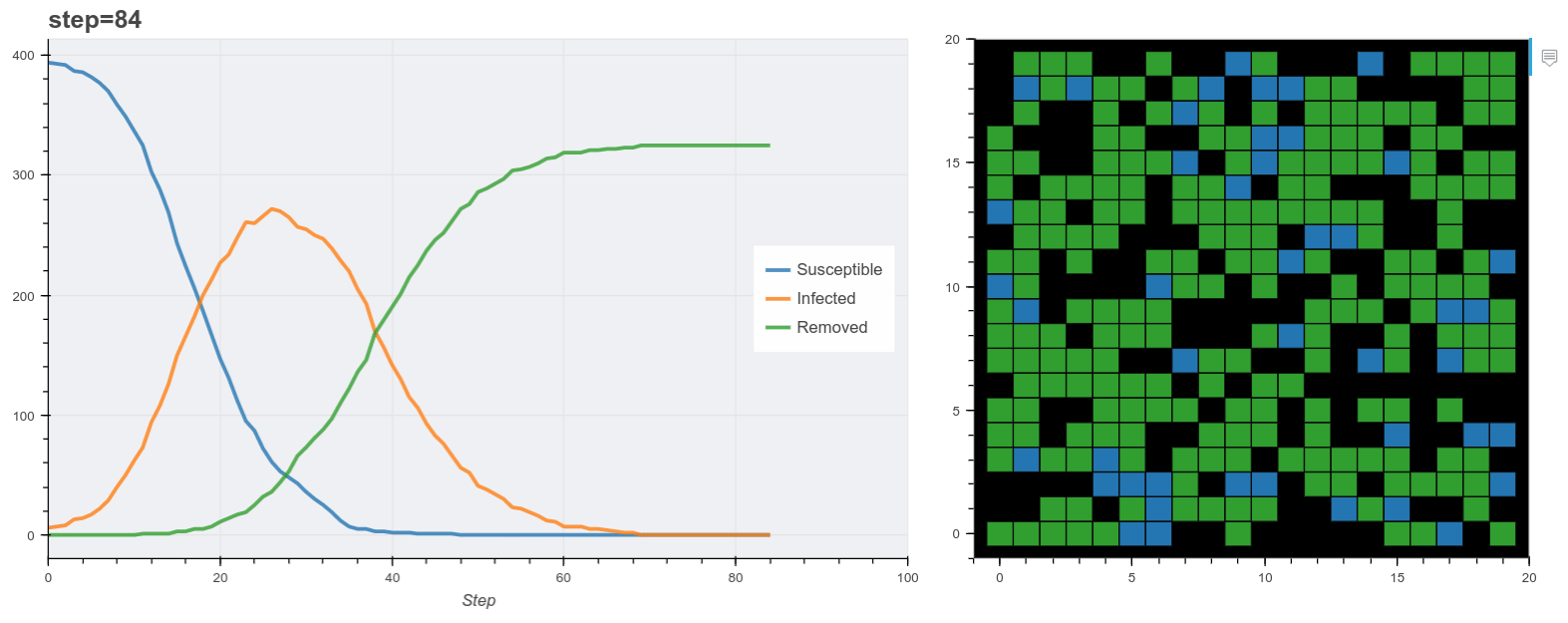
Finally we use these plot functions to show the model data as it runs. Mesa has it’s own visualization API also but this method is useful for running the model inside a Jupyter notebook. Here I used Panel to make two bokeh panes and update them at each step. (You don’t have to use Panel for this).
import panel as pn
pn.extension()
plot_pane = pn.pane.Bokeh()
grid_pane = pn.pane.Bokeh()
pn.Row(plot_pane,grid_pane,sizing_mode='stretch_width')
steps=100
pop=400
model = InfectionModel(pop, 20, 20, ptrans=0.25, death_rate=0.01)
for i in range(steps):
model.step()
p1=plot_states_bokeh(model,title='step=%s' %i)
plot_pane.object = p1
p2=plot_cells_bokeh(model)
grid_pane.object = p2
time.sleep(0.2)
The final output is shown below. You’ll notice the simulation reproduces the general pattern of states transitions of the SIR model. Beyond that, this is obviously a very simple model. For example most real world models normally probably don’t use grids like this, rather networks of contacts. A more realistic synthetic populaiton would describe additional features of each agent like age, household status and so on derived from demographic data of the target country.
References
- S. Venkatramanan, B. Lewis, J. Chen, D. Higdon, A. Vullikanti, and M. Marathe, “Using data-driven agent-based models for forecasting emerging infectious diseases,” Epidemics, vol. 22, pp. 43–49, 2018.