Using Molecular Nodes in Blender to visualise proteins
Background
Blender is a free program capable of advanced 3D modelling, animation and rendering. It is used to create photo realistic images for many applications such as animated films or architecture. I used to be interested in using Blender to create images of proteins and other molecules. This was a rather convoluted process. It seems things have moved on since I last looked at this area. There is now a very useful addon called Molecular Nodes that can import and render pdb structures all in one using geometry nodes. This is a way of applying a set of geometric transformations to objects like meshes. They can be combined to create new complex structures. This means you can import a pdb file as a basic set of points and apply these nodes to make visualisations.
Molecular Nodes
Like most 3D modelling software, Blender is ‘easy to learn but difficult to master’. So applying geometry nodes yourself to molecular structures would be hard for beginners. Molecular Nodes is an addon written and maintained by structural biologist Brady Johnston. It handles much of this process for you. No tutorial is given here as the instructions are well detailed by the author on his youtube channel. Just follow along with the examples. Though the version you use might be somewhat different to the one used in the tutorials. Installation is easy. Just download the zip file from the release page and install inside Blender via Preferences->Add ons. Here are a couple of basic examples of what you can do.
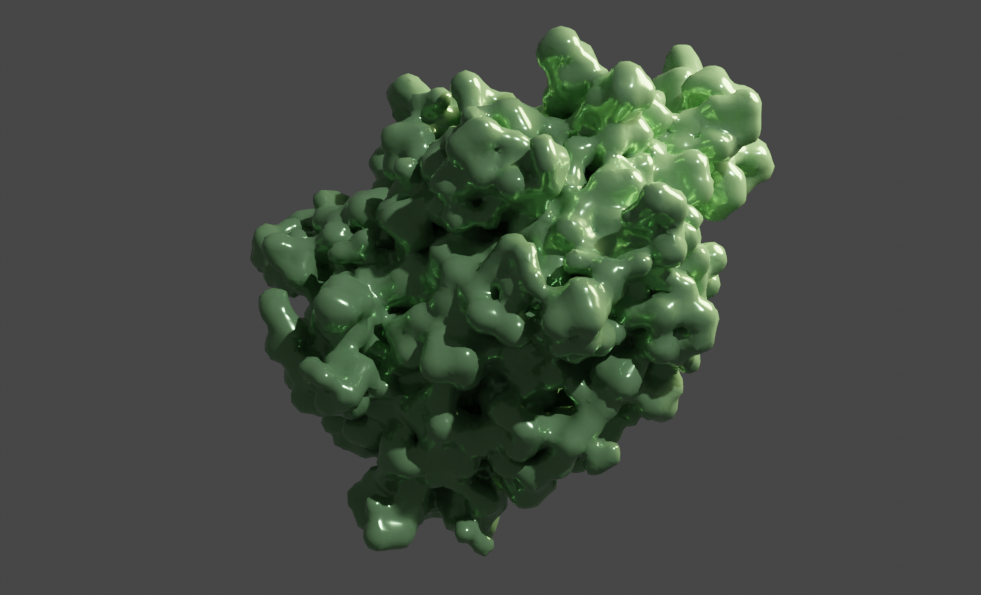
Fatty-acid binding protein
This example is the NMR structure of a fatty-acid binding protein. These proteins carry fatty acids and other lipids in the cellular environment, and are thus involved in processes such as FA uptake, transport, and oxidation.
NMR data comes with multiple states which can be animated using Molecular Nodes. This is quite straightforward (certainly compared to doing it yourself). Here I just added a glossy material and rotated the object as well. This gives an impression of the flexibility of the surface structures of the protein as the side chains move in solution:
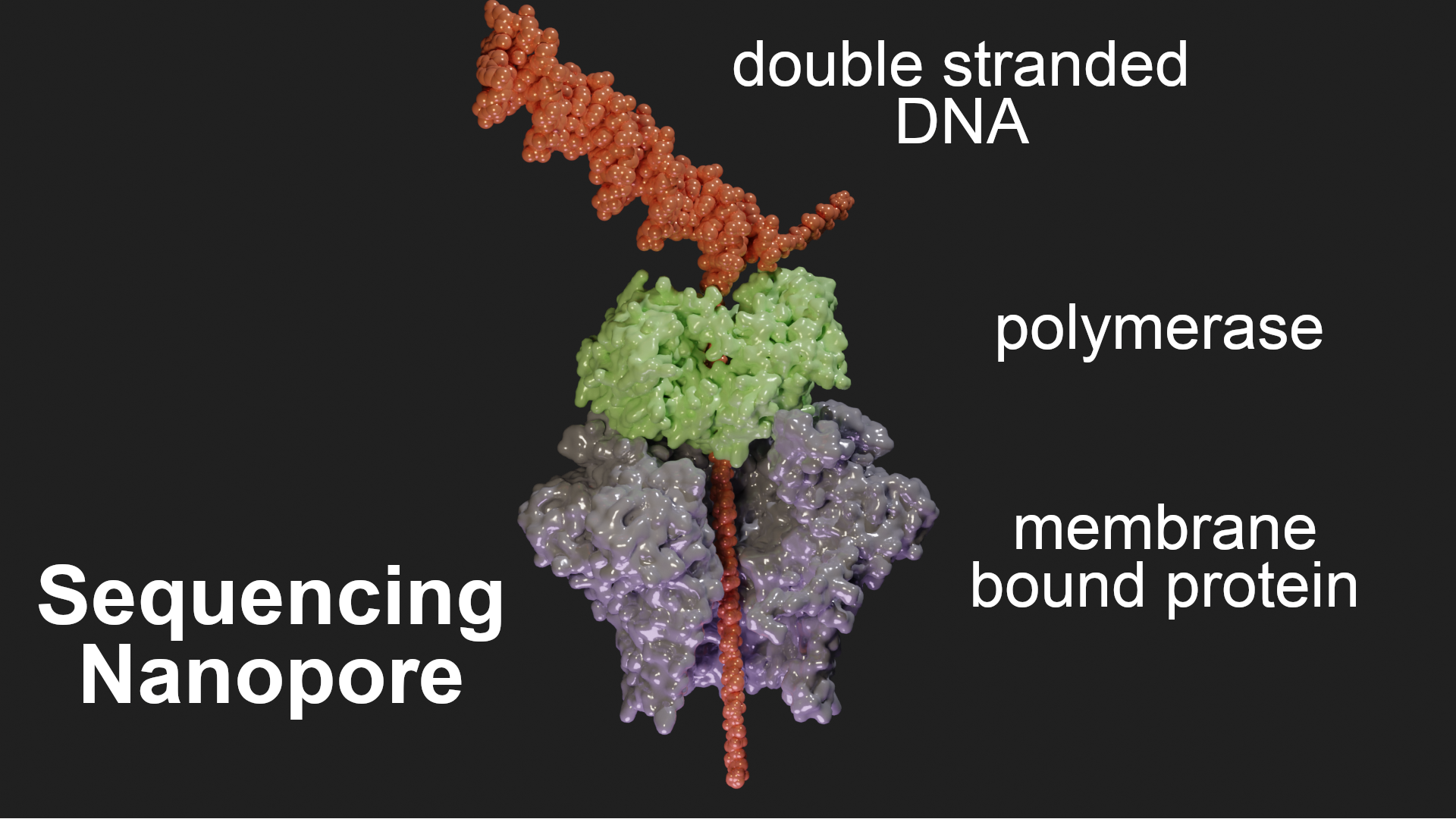
DNA sequencing nanopore
Nanopore sequencers are composed of nanopores embedded in a membrane that splits a salt solution into two chambers. A voltage is applied across the membrane, which causes ionic flow that can be measured. When a DNA strand is pulled through the pore, the ion flow is partially blocked, leading to a reduction in the observed current. The four types of nucleic acid bases are each associated with a different level of ion current change, which enables their identification. To visualise this I used the structures detailed in the pdb Molecule of the Month page. I imported the individual structures with the addon and rendered them in surface style. The DNA molecule I artificially made in Pymol (using the fnab command) and imported also. You can make DNA with the addon but I didn’t know how to use it properly to show the DNA unwinding as it goes into the pore. This is a hack and not that realistic but it’s mainly about making it look clear.
Shown below, the membrane protein is where the DNA passing through can be sensed. The polymerase (in green) slows down the DNA threading to make the signals more readable in real time. I also removed some chains from the membrane protein using the selection->chain node to show the pore more clearly.