Sequence alignment viewer with Qt/PySide2
Background
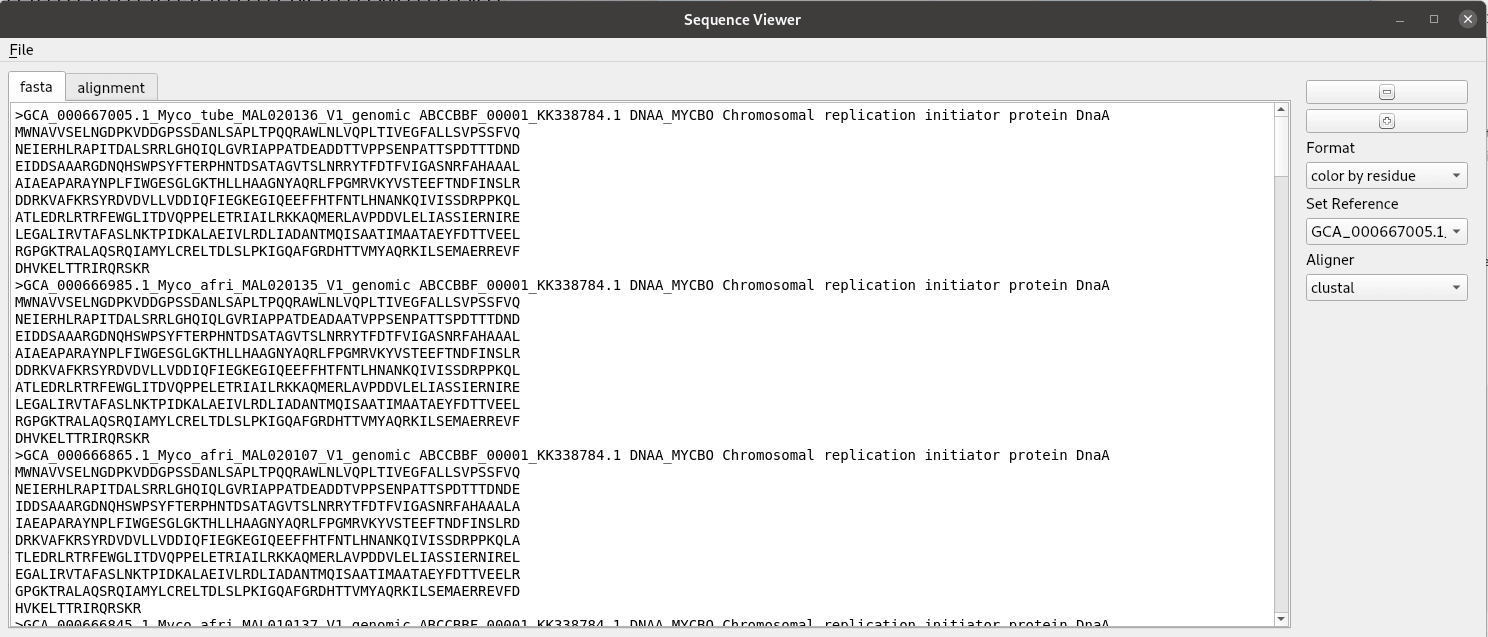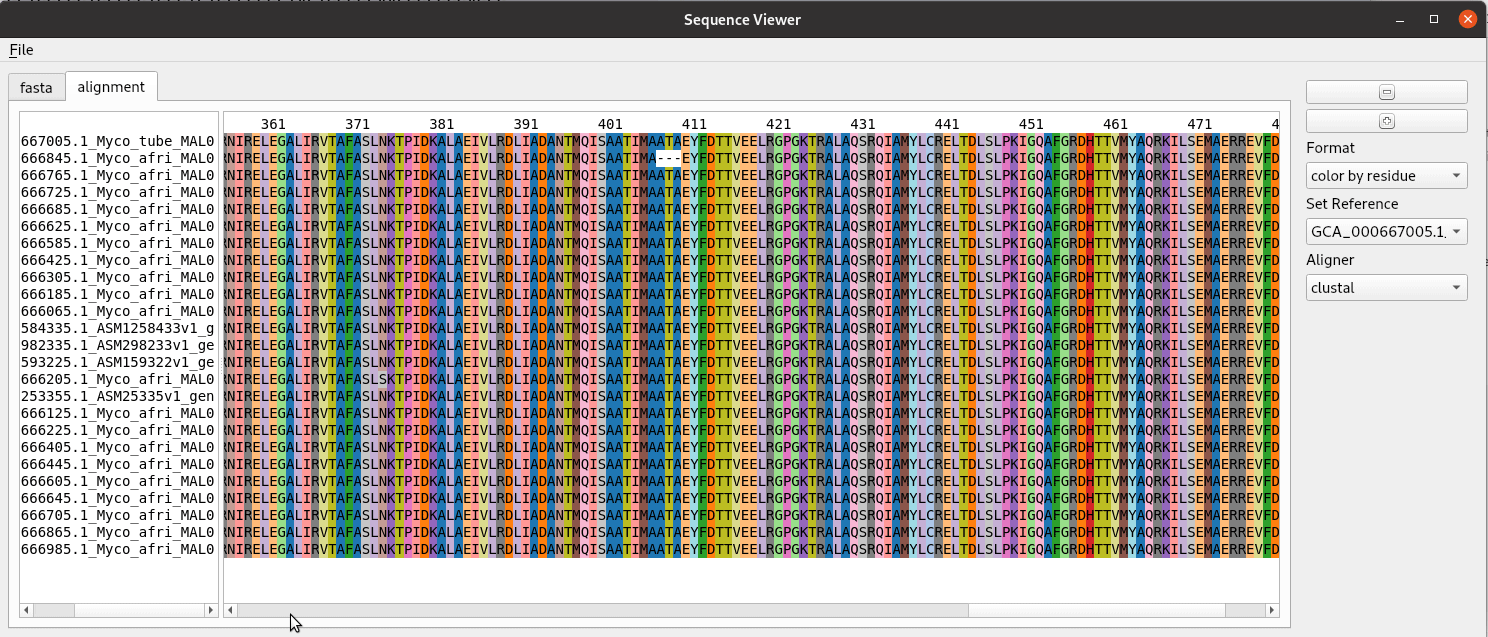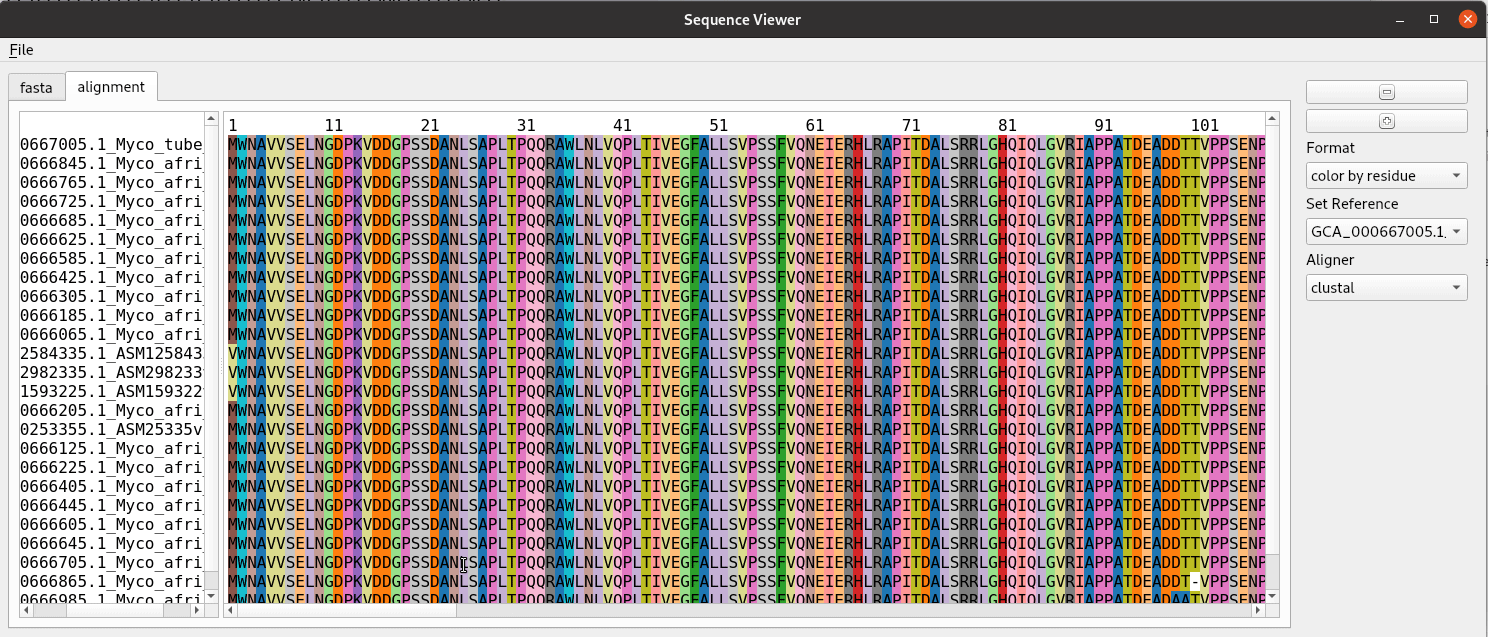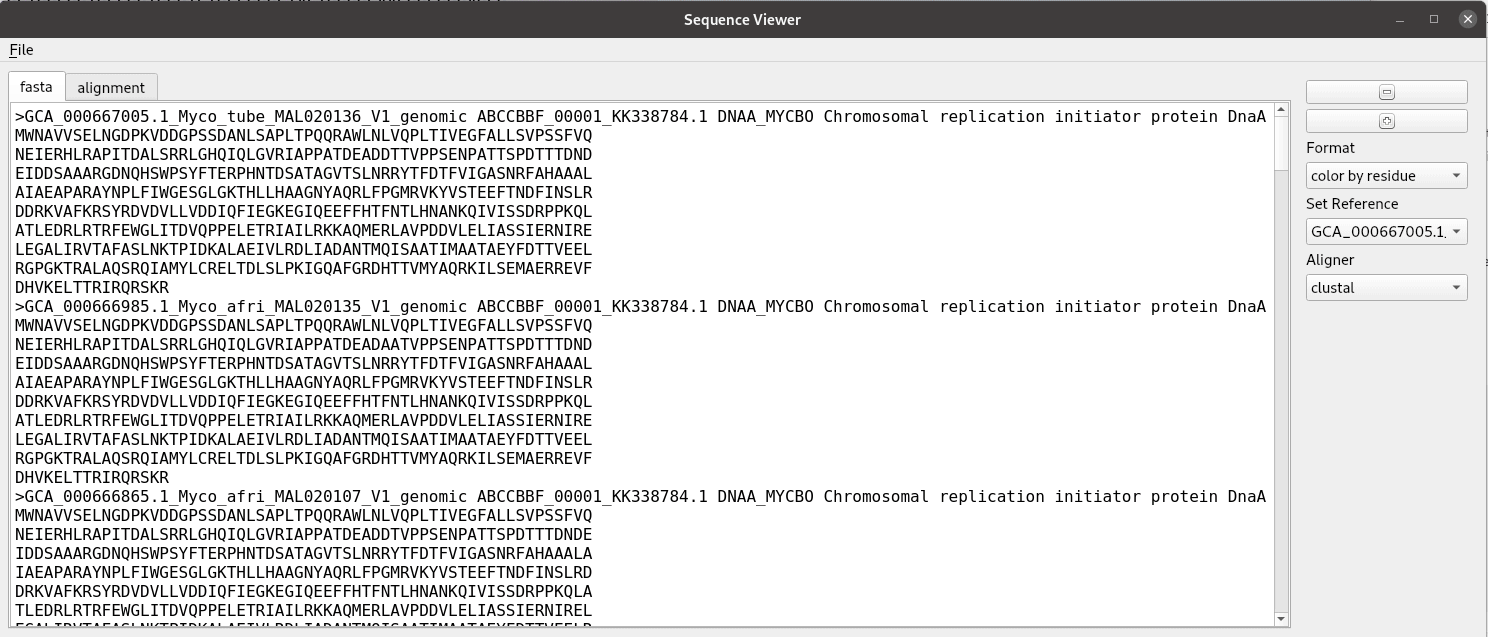
Title speaks for itself here. A sequence alignment is made here from a fasta file with clustal and read in using BioPython to a list of SeqRecord objects. The rest is just making a graphical interface for showing the alignment. We use a custom QPlainTextEdit class for the text displays. The AlignmentWidget class contains a QSplitter with areas for the labels and sequence separated. The draw_alignment method handles the drawing and coloring of the sequence alignment. The fastest way to do this is add the text letter by letter as html with the color specified. Despite it’s name QPlainTextEdit can handle html. This widget was developed for use in the pathogenie tool.
What’s missing
Missing is a way to make the two parts of the alignment scroll vertically together. These are in a QSplitter and need to use a shared scrollbar. Also needed is a way to set a reference sequence from one of the entries so that we can color
Imports
from PySide2 import QtCore, QtGui
from PySide2.QtCore import QObject
from PySide2.QtWidgets import *
from PySide2.QtGui import *
import sys, os, io
import numpy as np
import pandas as pd
import string
from Bio import SeqIO
def clustal_alignment(filename=None, seqs=None, command="clustalw"):
"""Align sequences with clustal"""
if filename == None:
filename = 'temp.faa'
SeqIO.write(seqs, filename, "fasta")
name = os.path.splitext(filename)[0]
command = get_cmd('clustalw')
from Bio.Align.Applications import ClustalwCommandline
cline = ClustalwCommandline(command, infile=filename)
stdout, stderr = cline()
align = AlignIO.read(name+'.aln', 'clustal')
return align
class PlainTextEditor(QPlainTextEdit):
def __init__(self, parent=None, **kwargs):
super(PlainTextEditor, self).__init__(parent, **kwargs)
font = QFont("Monospace")
font.setPointSize(10)
font.setStyleHint(QFont.TypeWriter)
self.setFont(font)
return
def zoom(self, delta):
if delta < 0:
self.zoomOut(1)
elif delta > 0:
self.zoomIn(1)
def contextMenuEvent(self, event):
menu = QMenu(self)
copyAction = menu.addAction("Copy")
clearAction = menu.addAction("Clear")
zoominAction = menu.addAction("Zoom In")
zoomoutAction = menu.addAction("Zoom Out")
action = menu.exec_(self.mapToGlobal(event.pos()))
if action == copyAction:
self.copy()
elif action == clearAction:
self.clear()
elif action == zoominAction:
self.zoom(1)
elif action == zoomoutAction:
self.zoom(-1)
class AlignmentWidget(QWidget):
"""Widget for showing sequence alignments"""
def __init__(self, parent=None):
super(AlignmentWidget, self).__init__(parent)
l = QHBoxLayout(self)
self.setLayout(l)
self.m = QSplitter(self)
l.addWidget(self.m)
self.left = PlainTextEditor(self.m, readOnly=True)
self.right = PlainTextEditor(self.m, readOnly=True)
self.left.setLineWrapMode(QPlainTextEdit.NoWrap)
self.right.setLineWrapMode(QPlainTextEdit.NoWrap)
self.m.setSizes([200,300])
self.m.setStretchFactor(1,2)
return
class SequencesViewer(QMainWindow):
"""Viewer for sequences and alignments"""
def __init__(self, parent=None, filename=None, title='Sequence Viewer'):
super(SequencesViewer, self).__init__(parent)
self.setWindowTitle(title)
self.setAttribute(QtCore.Qt.WA_DeleteOnClose)
self.setGeometry(QtCore.QRect(200, 200, 1000, 600))
self.setMinimumHeight(150)
self.recs = None
self.aln = None
self.add_widgets()
self.show()
return
def add_widgets(self):
"""Add widgets"""
self.main = QWidget(self)
self.setCentralWidget(self.main)
l = QHBoxLayout(self.main)
self.main.setLayout(l)
self.tabs = QTabWidget(self.main)
l.addWidget(self.tabs)
self.ed = ed = PlainTextEditor(self, readOnly=True)
self.ed.setLineWrapMode(QPlainTextEdit.NoWrap)
self.tabs.addTab(self.ed, 'fasta')
self.alnview = AlignmentWidget(self.main)
self.tabs.addTab(self.alnview, 'alignment')
sidebar = QWidget()
sidebar.setFixedWidth(180)
l.addWidget(sidebar)
l2 = QVBoxLayout(sidebar)
l2.setSpacing(5)
l2.setAlignment(QtCore.Qt.AlignTop)
btn = QPushButton()
btn.clicked.connect(self.zoom_out)
iconw = QIcon.fromTheme('zoom-out')
btn.setIcon(QIcon(iconw))
l2.addWidget(btn)
btn = QPushButton()
btn.clicked.connect(self.zoom_in)
iconw = QIcon.fromTheme('zoom-in')
btn.setIcon(QIcon(iconw))
l2.addWidget(btn)
lbl = QLabel('Format')
l2.addWidget(lbl)
w = QComboBox()
w.addItems(['no color','color by residue','color by difference'])
w.setCurrentIndex(1)
w.activated.connect(self.show_alignment)
self.formatchoice = w
l2.addWidget(w)
lbl = QLabel('Set Reference')
l2.addWidget(lbl)
w = QComboBox()
w.activated.connect(self.set_reference)
self.referencechoice = w
l2.addWidget(w)
lbl = QLabel('Aligner')
l2.addWidget(lbl)
w = QComboBox()
w.setCurrentIndex(1)
w.addItems(['clustal','muscle'])
self.alignerchoice = w
l2.addWidget(w)
self.create_menu()
return
def create_menu(self):
"""Create the menu bar for the application. """
self.file_menu = QMenu('&File', self)
self.menuBar().addMenu(self.file_menu)
self.file_menu.addAction('&Load Fasta File', self.load_fasta,
QtCore.Qt.CTRL + QtCore.Qt.Key_F)
self.file_menu.addAction('&Save Alignment', self.save_alignment,
QtCore.Qt.CTRL + QtCore.Qt.Key_S)
return
def scroll_top(self):
vScrollBar = self.ed.verticalScrollBar()
vScrollBar.triggerAction(QScrollBar.SliderToMinimum)
return
def zoom_out(self):
self.ed.zoom(-1)
self.alnview.left.zoom(-1)
self.alnview.right.zoom(-1)
return
def zoom_in(self):
self.ed.zoom(1)
self.alnview.left.zoom(1)
self.alnview.right.zoom(1)
return
def load_fasta(self, filename=None):
"""Load fasta file"""
if filename == None:
filename, _ = QFileDialog.getOpenFileName(self, 'Open File', './',
filter="Fasta Files(*.fa *.fna *.fasta);;All Files(*.*)")
if not filename:
return
recs = list(SeqIO.parse(filename, 'fasta'))
self.load_records(recs)
return
def load_records(self, recs):
"""Load seqrecords list"""
self.recs = recs
self.reference = self.recs[0]
rdict = SeqIO.to_dict(recs)
self.show_fasta()
self.show_alignment()
self.referencechoice.addItems(list(rdict.keys()))
return
def set_reference(self):
ref = self.referencechoice.currentText()
return
def show_fasta(self):
"""Show records as fasta"""
recs = self.recs
if recs == None:
return
self.ed.clear()
for rec in recs:
s = rec.format('fasta')
self.ed.insertPlainText(s)
self.scroll_top()
return
def align(self):
"""Align current sequences"""
if self.aln == None:
outfile = 'temp.fa'
SeqIO.write(self.recs, outfile, 'fasta')
self.aln = clustal_alignment(outfile)
return
def show_alignment(self):
format = self.formatchoice.currentText()
self.draw_alignment(format)
return
def draw_alignment(self, format='color by residue'):
"""Show alignment with colored columns"""
left = self.alnview.left
right = self.alnview.right
chunks=0
offset=0
diff=False
self.align()
aln = self.aln
left.clear()
right.clear()
self.scroll_top()
colors = tools.get_protein_colors()
format = QtGui.QTextCharFormat()
format.setBackground(QtGui.QBrush(QtGui.QColor('white')))
cursor = right.textCursor()
ref = aln[0]
l = len(aln[0])
n=60
s=[]
if chunks > 0:
chunks = [(i,i+n) for i in range(0, l, n)]
else:
chunks = [(0,l)]
for c in chunks:
start,end = c
lbls = np.arange(start+1,end+1,10)-offset
head = ''.join([('%-10s' %i) for i in lbls])
cursor.insertText(head)
right.insertPlainText('\n')
left.appendPlainText(' ')
for a in aln:
name = a.id
seq = a.seq[start:end]
left.appendPlainText(name)
line = ''
for aa in seq:
c = colors[aa]
line += '<span style="background-color:%s;">%s</span>' %(c,aa)
cursor.insertHtml(line)
right.insertPlainText('\n')
return
def save_alignment(self):
filters = "clustal files (*.aln);;All files (*.*)"
filename, _ = QFileDialog.getSaveFileName(self,"Save Alignment",
"",filters)
if not filename:
return
SeqIO.write(self.aln,filename,format='clustal')
return
Result
Running the app
Here is the code to make a script that will launch from the command line:
def main():
"Run the application"
import sys, os
from argparse import ArgumentParser
parser = ArgumentParser(description='sequence viewer tool')
parser.add_argument("-f", "--fasta", dest="filename",default=None,
help="input fasta file", metavar="FILE")
args = vars(parser.parse_args())
app = QApplication(sys.argv)
sv = widgets.SequencesViewer(**args)
if args['filename'] != None:
sv.load_fasta(args['filename'])
sv.show()
app.exec_()
if __name__ == '__main__':
main()